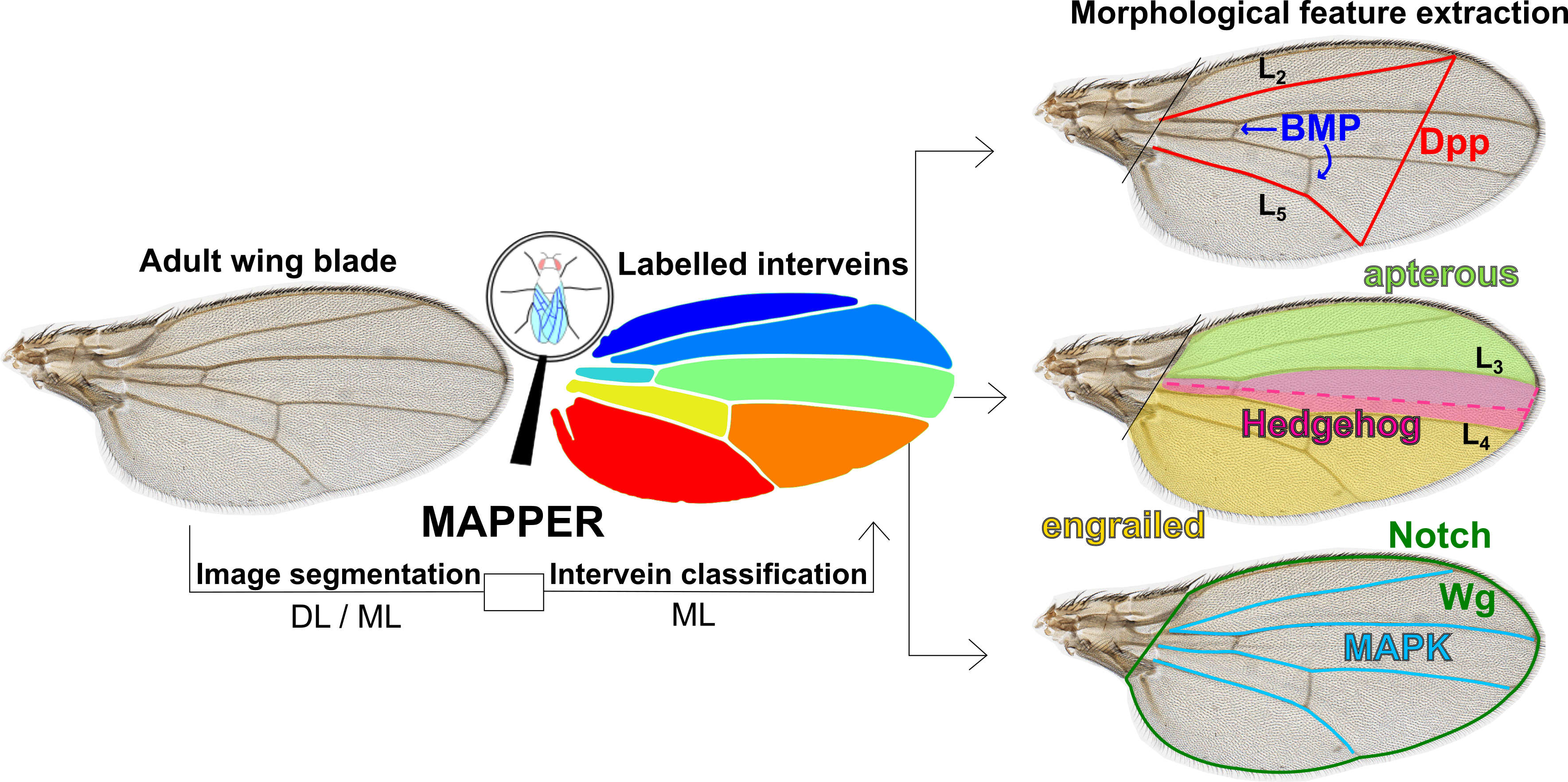
Here is a repository for the manuscript titled MAPPER: An open-source, high-dimensional image analysis pipeline unmasks differential regulation of Drosophila wing features developed and written by Nilay Kumar and Francisco Huizar in the Zartman lab at the University of Notre Dame. You can find the pre-print of the paper on bioRxiv here. The bulk of the code was built by Nilay Kumar and co-developed by Francisco Huizar. Dr. Ramezan Paravi Torghabeh and Dr. Pavel Brodskiy provided guidance for code development. Experimental work and validation was carried out by Nilay Kumar, Dr. Maria Unger, Trent Robinett, Keity J. Farfan-Pira, and Dharsan Soundarrajan. This work was done within the Multicellular Systems Engineering Lab at the University of Notre Dame and the Laboratory of Growth Biology and Morphogenesis at the Center for Research and Advanced Studies of the National Polytechnical Institute (Cinvestav). Please direct any questions to the principal investigator, Dr. Jeremiah Zartman.
All code for the MAPPER application was done using MATLAB.
Open-source license agreement
- This code is made publicly available for use under the GNU Lesser General Public License, version 2.1
- You can find the license agreement for MAPPER here
- We kindly ask users of the code and any derivative work to cite the original publication:
†Kumar, N., †Huizar, F.J., Farfán-Pira K.J., Brodskiy, P., Soundarrajan, D.S., Nahmad, M., Zartman, J.J.; MAPPER: An open-source, high-dimensional image analysis pipeline unmasks differential regulation of Drosophila wing features. Frontiers in Genetics (2022). https://doi.org/10.3389/fgene.2022.869719 † These authors contributed equally.
The below licensing statements are verbatim statements from the Free and Open Source Software Auditing (FOSSA) team originating from this webpage
Users of this code must:
- Include a copy of the full license text and the original copyright notice
- Make available the source code when you distribute a derivative work based on the licensed library
- License any derivative works of the library under the same or later version of the LGPL or GPL
The LGPL license allows users of the licensed code to:
- Use the code commercially: Like GPL, LGPL imposes no conditions on using the code in software that’s sold commercially
- Change the code: Users can rework the code, but if they distribute these modifications, they must release these updates in source code form
- Place warranty: Distributors of the original code can offer their own warranty on the licensed software
Instructions to run the application
- You can download all of the associated code here
- Once you have downloaded the .ZIP folder, extract the folder to an easily accessible location
- You can download the full user manual for how to begin using MAPPER here
- Alternatively, the full user manual is available within the downloadable MAPPER folder
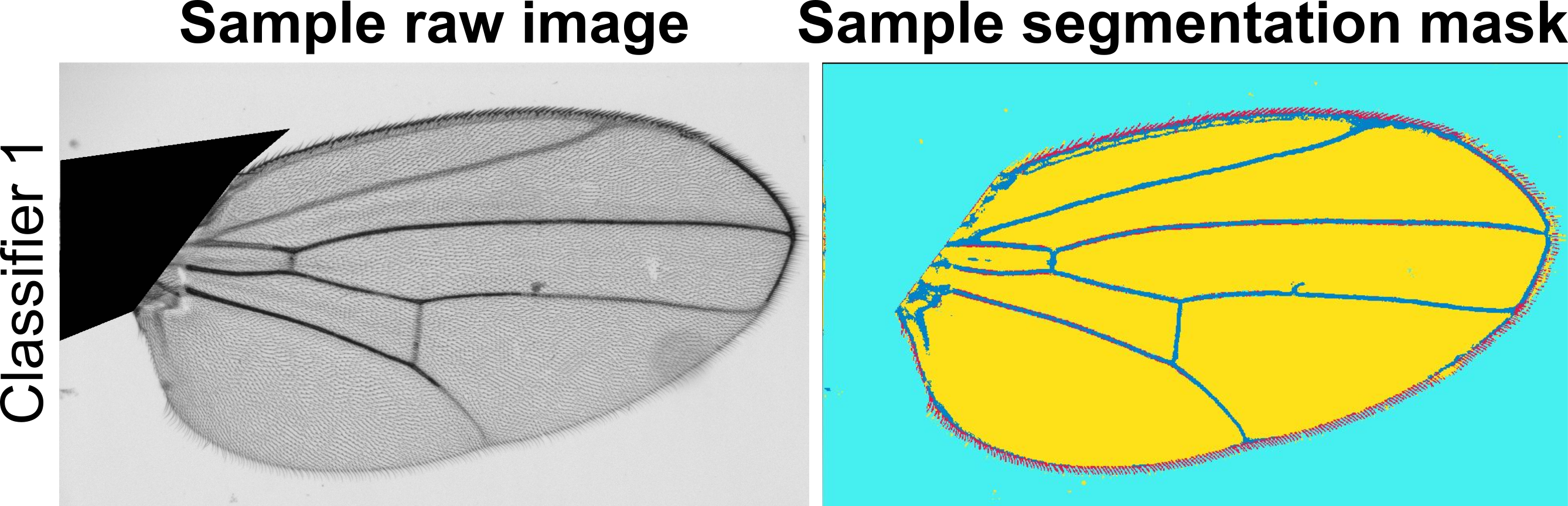
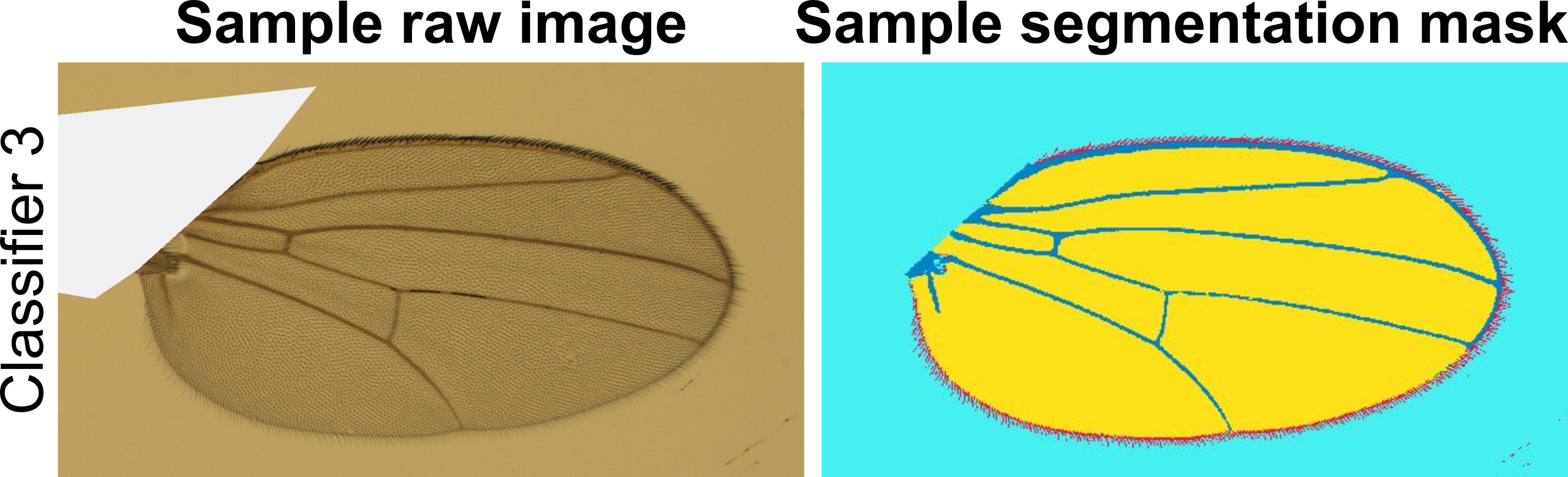
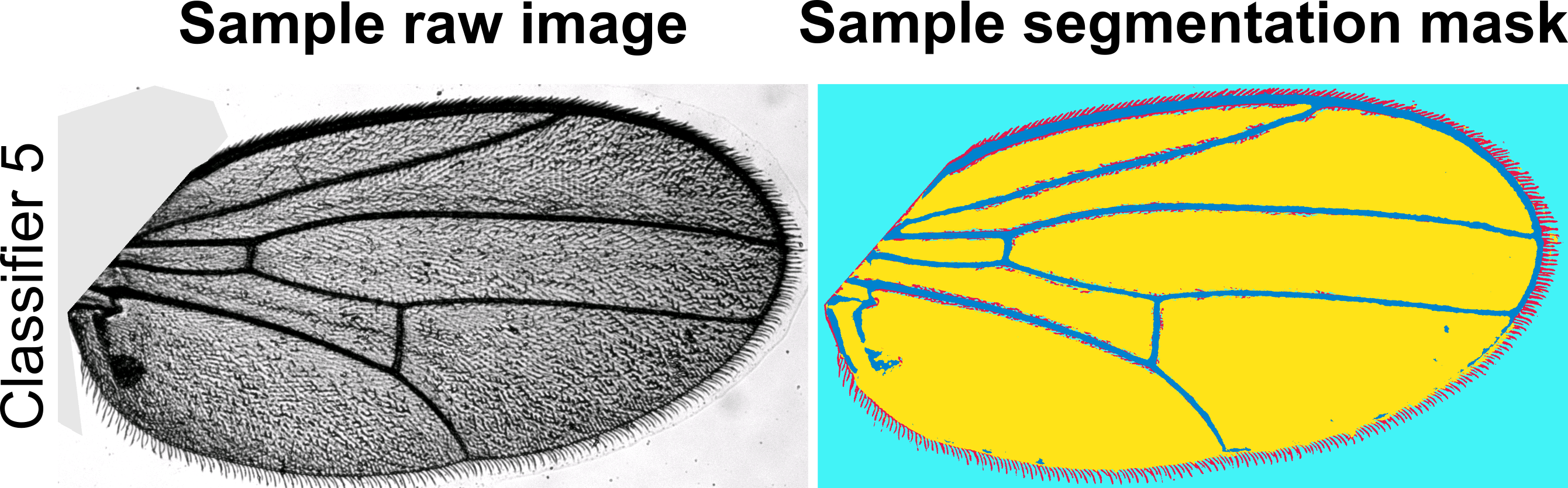
Available ILASTIK pixel classification modules
Below you will find pre-trained pixel classification modules in ILASTIK for several wing images we have already processed. These modules are crucial for step five of the MAPPER user manual. Below each module link, you will find a representative image of the Drosophila wings that were used to train the module. You should download and use the ILASTIK module that has the closest resemblance in lighting, background, brightness, contrast, and saturation to the images you would like to process. If none of the available ILASTIK modules closely resemble the images you would like to process, there are detailed instructions in the user manual on how to train your own ILASTIK module. NOTE: The number of channels of your images must match the number of channels in the training data for the ILASTIK module you choose (i.e., RGB channel images must have an ILASTIK module trained on RGB channel images).
- Download ILASTIK module 1
- Source: Nilay Kumar, Zartman Lab, Univeristy of Notre Dame
- One channel (Grayscale)
- Download ILASTIK module 2
- Source: Nilay Kumar, Zartman Lab, Univeristy of Notre Dame
- One channel (Grayscale)

- Download ILASTIK module 3
- Source: Nilay Kumar, Zartman Lab, Univeristy of Notre Dame
- Three channels (RGB)
- Download ILASTIK module 4
- Source: Nilay Kumar, Zartman Lab, Univeristy of Notre Dame
- One channel (Grayscale)

- Download ILASTIK module 5
- Source: Nilay Kumar, Zartman Lab, Univeristy of Notre Dame
- One channel (Grayscale)
- Download ILASTIK module 6
- Source: Nilay Kumar, Zartman Lab, Univeristy of Notre Dame
- Three channels (RGB)

U-Net deep learning trained model
- Supplementary File 1 sections S1 and S2 of the manuscript references a U-Net trained model that is linked here
- The full folder contains:
- Image training data in the “imagedata” subfolder
- The python code for training the U-net in an iPython notebook labeled “Deep learning-Unet-CrossEntropy-Four_classes”
- The python code for using the U-net in an iPython notebook labeled “Model Usage”
- A presentation on utilizing U-Net for Drosophila wing segmentation in a Microsoft Powerpoint labeled “wingSegmentation”
- For training the intervein classifier, MATLAB’s image labeler app was used and the training resources can be found here
- More information for this is explained in section S2 of Supplementary File 1
Supplementary File 2 R Notebook
- Figures 3 and 6 of the main text reference a supplementary file for statistical calculations
- The supplementary file is in the form of an R Notebook and can be found here
- Plots generated from the notebook were exported as SVG files and modified in Inkscape vector graphics software for final versions of the manuscript plots
Raw Data Sheets
- Data for Figures 3, 6, S7, and S8 can be found here. Explicit operations on the data sheets are found within Supplementary File 2
- Data for Figures 4, 5, S9, and S10 can be found here for wing features and here for EFD coefficients
- Data for Figures 7, S11, and S12 can be found here
Acknowledgements
We would like to thank the South Bend Medical Foundation for generous access to their Apero Slide Scanner. We would like to thank Dr. Ramezan Paravi Torghabeh, Vijay Kumar Naidu Velagala, Dr. Megan Levis, and Dr. Qinfeng Wu for technical assistance and scientific discussions related to the project. The work in this manuscript was supported in part by NIH Grant R35GM124935, NSF award CBET-1553826, NSF-Simons Pilot award through Northwestern University, the Notre Dame International Mexico Faculty Grant Program, and grant CB-014-01-236685 from the Concejo Nacional de Ciencia y Tecnología of Mexico.